Extending Python with C or C++
Python has an elegant interface to C and C++ which allows you to write the most computation-intensive parts of your code in a low-level language with excellent performance. The following example is due to Matthew Fowler who shared his code on github.
The following example extends Python with a simple program for computing the
standard deviation of many small matrices. While numpy is optimized for large
matrices, it runs slower for millions of smaller calculations. These can be more
efficiently computed with a simple C++ program. In this exercise we will write
this code in stddev.cpp below.
The following code requires the use of Python.h which supplies a number of
types required to convert Python objects into C objects. This procedure is
described at length in the
documentation.
Components such as PyObject, PyArg_ParseTuple, and PyMethodDef are all
part of this interface.
In [1]:
%%file stddev.cpp
#include <Python.h>
#include <vector>
#include <numeric>
#include <iterator>
// Example Python vs C comparison
// via https://github.com/mattfowler/PythonCExtensions
// https://medium.com/coding-with-clarity/
// speeding-up-python-and-numpy-c-ing-the-way-3b9658ed78f4
double standardDeviation(std::vector<double> v)
{
double sum = std::accumulate(v.begin(), v.end(), 1.0);
double mean = sum / v.size();
double squareSum = std::inner_product(
v.begin(), v.end(), v.begin(), 0.0);
return sqrt(squareSum / v.size() - mean * mean);
}
static PyObject * std_standard_dev(PyObject *self, PyObject* args)
{
PyObject* input;
PyArg_ParseTuple(args, "O", &input);
int size = PyList_Size(input);
std::vector<double> list;
list.resize(size);
for(int i = 0; i < size; i++) {
list[i] = PyFloat_AS_DOUBLE(PyList_GET_ITEM(input, i));
}
return PyFloat_FromDouble(standardDeviation(list));
}
static PyMethodDef std_methods[] = {
{"standard_dev", std_standard_dev,METH_VARARGS,
"YYY Return the standard deviation of a list. XX"},
{NULL,NULL} /* sentinel */
};
static struct PyModuleDef stdmodule = {
PyModuleDef_HEAD_INIT,
"std", /* name of module */
NULL, /* module documentation, may be NULL */
-1,
std_methods
};
//x
PyMODINIT_FUNC PyInit_std(void)
{
return PyModule_Create(&stdmodule);
}
int main(int argc, char **argv)
{
wchar_t *program = Py_DecodeLocale(argv[0], NULL);
if (program == NULL) {
fprintf(stderr, "Fatal error: cannot decode argv[0]\n");
exit(1);
}
/* Add a built-in module, before Py_Initialize */
PyImport_AppendInittab("std", PyInit_std);
/* Pass argv[0] to the Python interpreter */
Py_SetProgramName(program);
/* Initialize the Python interpreter. Required. */
Py_Initialize();
PyMem_RawFree(program);
return 0;
}Overwriting stddev.cpp
The following Python code uses
distutils to package the
code from stdev.cpp into a module called std. We install the program below.
In [2]:
%%file setup.py
from distutils.core import setup, Extension
std_module = Extension('std', sources=['stddev.cpp'])
setup(name='std_performance',
version='1.0',
description='Module for calculating standard deviation.',
install_requires = ["cycler==0.10.0",
"kiwisolver==1.0.1"
"matplotlib==3.0.2",
"numpy==1.16.1",
"pandas==0.24.1",
"pyparsing==2.3.1",
"python-dateutil==2.8.0",
"pytz==2018.9",
"six==1.12.0"],
ext_modules=[std_module])Overwriting setup.py
In [3]:
# install this module in our local Python environment
! python setup.py install/Users/rpb/worker/dev/factory-bleed/conda/envs/dev/lib/python3.7/distutils/dist.py:274: UserWarning: Unknown distribution option: 'install_requires'
warnings.warn(msg)
running install
running build
running build_ext
building 'std' extension
x86_64-apple-darwin13.4.0-clang -fno-strict-aliasing -Wsign-compare -Wunreachable-code -DNDEBUG -fwrapv -O3 -Wall -Wstrict-prototypes -march=core2 -mtune=haswell -mssse3 -ftree-vectorize -fPIC -fPIE -fstack-protector-strong -O3 -pipe -fdebug-prefix-map=${SRC_DIR}=/usr/local/src/conda/${PKG_NAME}-${PKG_VERSION} -fdebug-prefix-map=/Users/rpb/worker/dev/factory-bleed/conda/envs/dev=/usr/local/src/conda-prefix -flto -Wl,-export_dynamic -march=core2 -mtune=haswell -mssse3 -ftree-vectorize -fPIC -fPIE -fstack-protector-strong -O3 -march=core2 -mtune=haswell -mssse3 -ftree-vectorize -fPIC -fPIE -fstack-protector-strong -O2 -pipe -D_FORTIFY_SOURCE=2 -mmacosx-version-min=10.9 -I/Users/rpb/worker/dev/factory-bleed/conda/envs/dev/include/python3.7m -c stddev.cpp -o build/temp.macosx-10.9-x86_64-3.7/stddev.o
clang-4.0: warning: -Wl,-export_dynamic: 'linker' input unused [-Wunused-command-line-argument]
x86_64-apple-darwin13.4.0-clang++ -bundle -undefined dynamic_lookup -Wl,-pie -Wl,-headerpad_max_install_names -Wl,-dead_strip_dylibs -Wl,-rpath,/Users/rpb/worker/dev/factory-bleed/conda/envs/dev/lib -L/Users/rpb/worker/dev/factory-bleed/conda/envs/dev/lib -flto -Wl,-export_dynamic -Wl,-pie -Wl,-headerpad_max_install_names -Wl,-dead_strip_dylibs -Wl,-rpath,/Users/rpb/worker/dev/factory-bleed/conda/envs/dev/lib -L/Users/rpb/worker/dev/factory-bleed/conda/envs/dev/lib -Wl,-pie -Wl,-headerpad_max_install_names -Wl,-dead_strip_dylibs -march=core2 -mtune=haswell -mssse3 -ftree-vectorize -fPIC -fPIE -fstack-protector-strong -O2 -pipe -D_FORTIFY_SOURCE=2 -mmacosx-version-min=10.9 -arch x86_64 build/temp.macosx-10.9-x86_64-3.7/stddev.o -o build/lib.macosx-10.9-x86_64-3.7/std.cpython-37m-darwin.so
ld: warning: -pie being ignored. It is only used when linking a main executable
ld: warning: ignoring file /Library/Developer/CommandLineTools/SDKs/MacOSX10.14.sdk/usr/lib/libSystem.tbd, file was built for unsupported file format ( 0x2D 0x2D 0x2D 0x20 0x21 0x74 0x61 0x70 0x69 0x2D 0x74 0x62 0x64 0x2D 0x76 0x33 ) which is not the architecture being linked (x86_64): /Library/Developer/CommandLineTools/SDKs/MacOSX10.14.sdk/usr/lib/libSystem.tbd
running install_lib
copying build/lib.macosx-10.9-x86_64-3.7/std.cpython-37m-darwin.so -> /Users/rpb/worker/dev/factory-bleed/conda/envs/dev/lib/python3.7/site-packages
running install_egg_info
Removing /Users/rpb/worker/dev/factory-bleed/conda/envs/dev/lib/python3.7/site-packages/std_performance-1.0-py3.7.egg-info
Writing /Users/rpb/worker/dev/factory-bleed/conda/envs/dev/lib/python3.7/site-packages/std_performance-1.0-py3.7.egg-info
Now that we have installed this extension module, we are free to use it in an example to determine the performance benefit.
In [4]:
import numpy as np
import random
import timeit
import pandas as pd
%matplotlib inline
from matplotlib import pyplot as plt
import std
lens = range(1000, 20000, 1000)
lp_time = []
py_time = []
np_time = []
c_time = []
for l in lens:
rands = [random.random() for _ in range(0, l)]
numpy_rands = np.array(rands)
np_time = np.append(np_time, timeit.timeit(lambda: np.std(numpy_rands), number=1000))
c_time = np.append(c_time, timeit.timeit(lambda: std.standard_dev(rands), number=1000))
data = np.array([np.transpose(np_time), np.transpose(c_time)])
df = pd.DataFrame(data.transpose(), index=lens, columns=['Numpy', 'C++'])
plt.figure()
df.plot()
plt.legend(loc='best')
plt.ylabel('Time (Seconds)')
plt.xlabel('Number of Elements')
plt.title('1k Runs of Standard Deviation')
plt.savefig('numpy_vs_c.png')
plt.show()<Figure size 432x288 with 0 Axes>
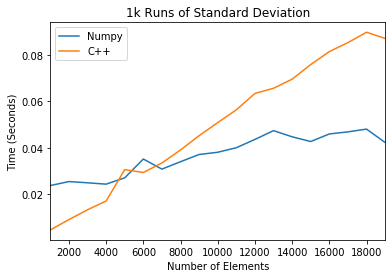
We find that this method outperforms numpy for small matrices.